
New members
Enrollment of new PhD students
We are happy to welcome Gustav, Aleksandra and Daniel in our lab! They just started their PhD journey embarking on exciting new projects.[2024-07-01]
Paper
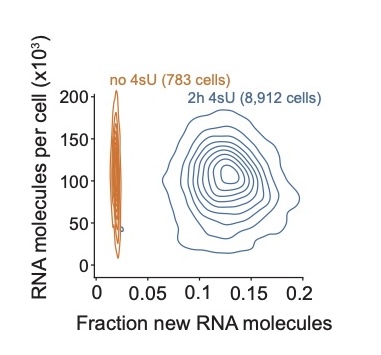
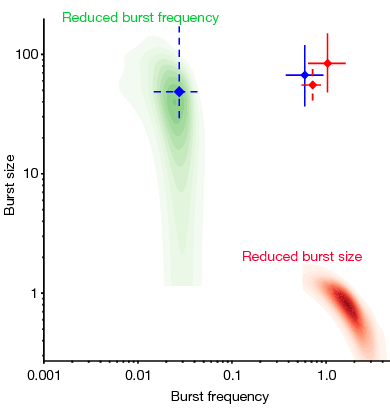
New insights into the dynamics of transcriptional bursting
In this paper we show that more precise transcriptional bursting inference is possible through temporal single-cell RNA-sequencing (using 4sU) that enabled us to demonstrate that bursts are typically equally long, irrespective of burst size, and that bursts that generate more RNA molecules have a higher synthesis rate. Congrats to the team for performing this revealing and fun project, that came out in Nature Cell Biology.[2024-06-22] [link]

Award
Torsten Söderberg Professorship 2022
Rickard was awarded the Torsten Söderbergs academy professorship 2022[2023-03-10] [link]
Paper
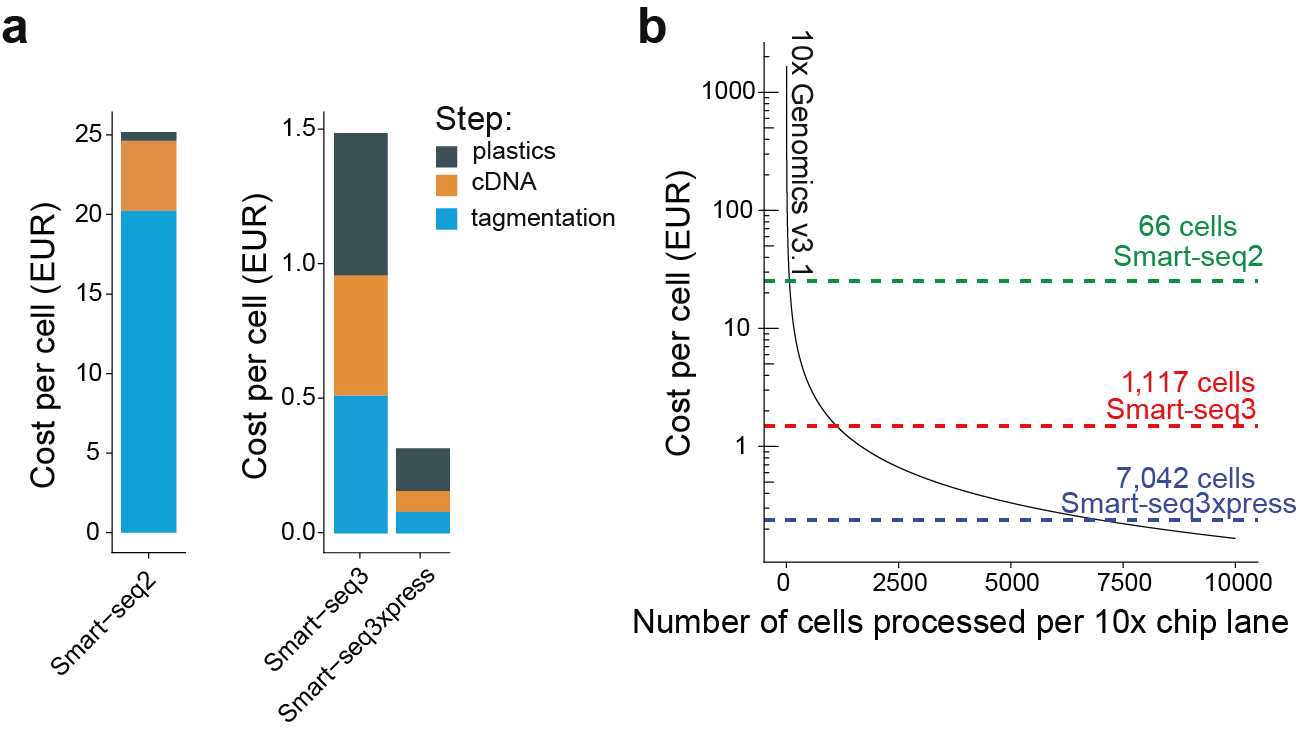
Smart-seq3xpress: Expressway to high-quality single-cell RNA-seq data
We are extremely happy to share our state-of-the-art plate-based method that is cost-efficient, scalable and sensitive, that cost per cell is now down to a whopping 0.25 EUR with costs per experiments mirroring 10x Genomics libraries. Just out in Nature Biotechnology.[2022-05-20] [link]
Paper
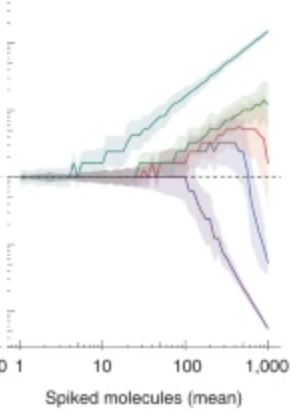
Molecular spikes
Our recent work on improving RNA counting in single cells using molecular spikes is just out in Nature Methods.[2022-04-25] [link]
Paper
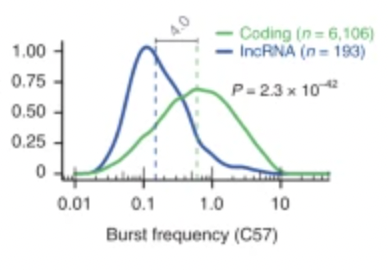
Transcriptional kinetics and molecular functions of long noncoding RNAs
In-depth characterization of lincRNAs expression in terms of transcriptional dynamics and demonstration of how single-cell data can be used to predict lincRNA function. Now out in Nature Genetics![2022-03-10] [link]

Paper
BAMboozle: the removal of human sequence variation for open sharing
Are you also disappointed not being able to share your nice (human) sequencing data because of donor's genetic privacy? We wrote a lightweight software to anonymize reads stored in regular .bam files! Available on biorxiv and tool is here: https://github.com/sandberg-lab/dataprivacy[2021-01-10] [link]
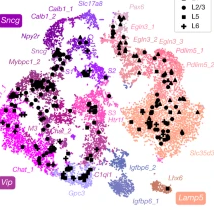
Paper
Multi-modal analysis of neurons using Patch-seq
Massive efforts to monitor electrophysiology, morphology, and transcriptomes in over 1,000 cells. We found that, although broad families of transcriptomic types had distinct morpho-electric phenotypes, individual transcriptomic types within the same family were not well separated in the morpho-electric space. All transcriptomics made with Smart-seq2 all mastered by Leo! Published in Nature.[2020-11-10] [link]
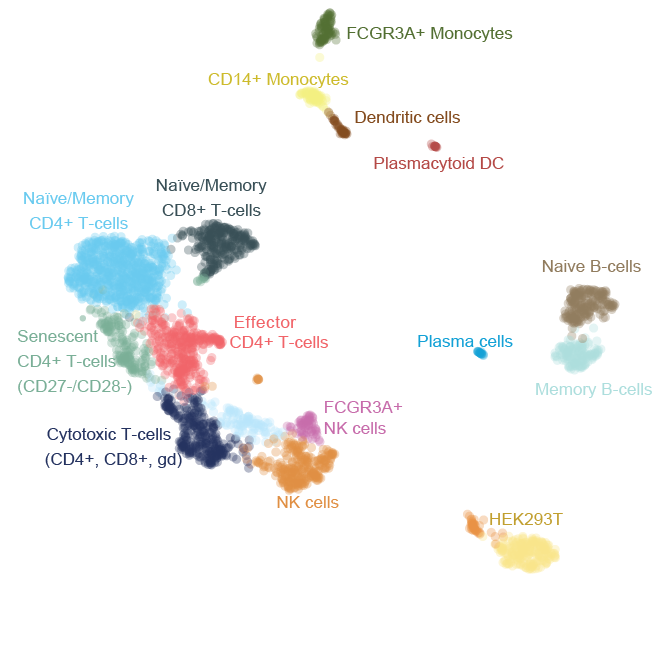
Paper
RNA counting and molecule reconstrution with Smart-seq3
scRNA-seq with improved sensitivity and unique abilities to reconstruct counted RNA molecules was published in nature biotechnology![2020-04-01] [link]
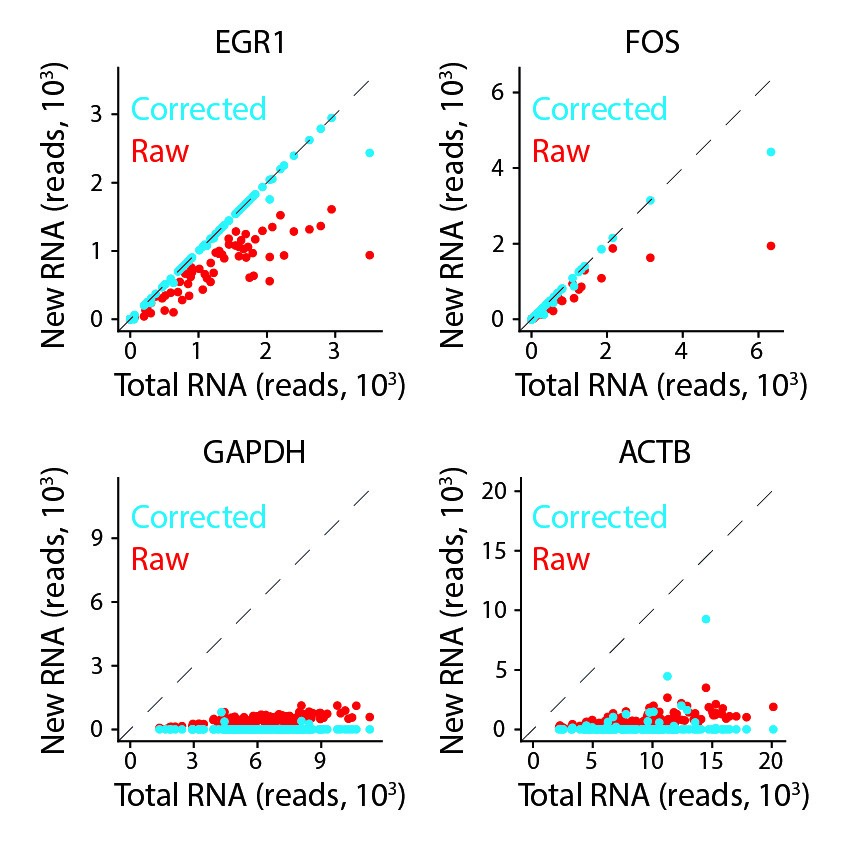
Paper
Single-cell RNA-sequencing of newly transcribed and pre-existing RNA
In collaboration with Patrick Cramers lab, we developed metabolic labelling for single-cell transcriptomics. Important for ongoing studies in the lab focusing on transcriptional dynamics. Published in Nature Communications.[2019-07-01] [link]
Paper
Genomic encoding of transcriptional burst kinetics
Transcription occurs in bursts. In this paper, we used single-cell RNA-sequencing with allelic resolution to infer the kinetics of transcription for thousands of genes. We found a transcriptome-wide role of enhancers in dictating burst frequencies, whereas core promoter elements affected burst size. Published in Nature.[2019-01-01] [link]
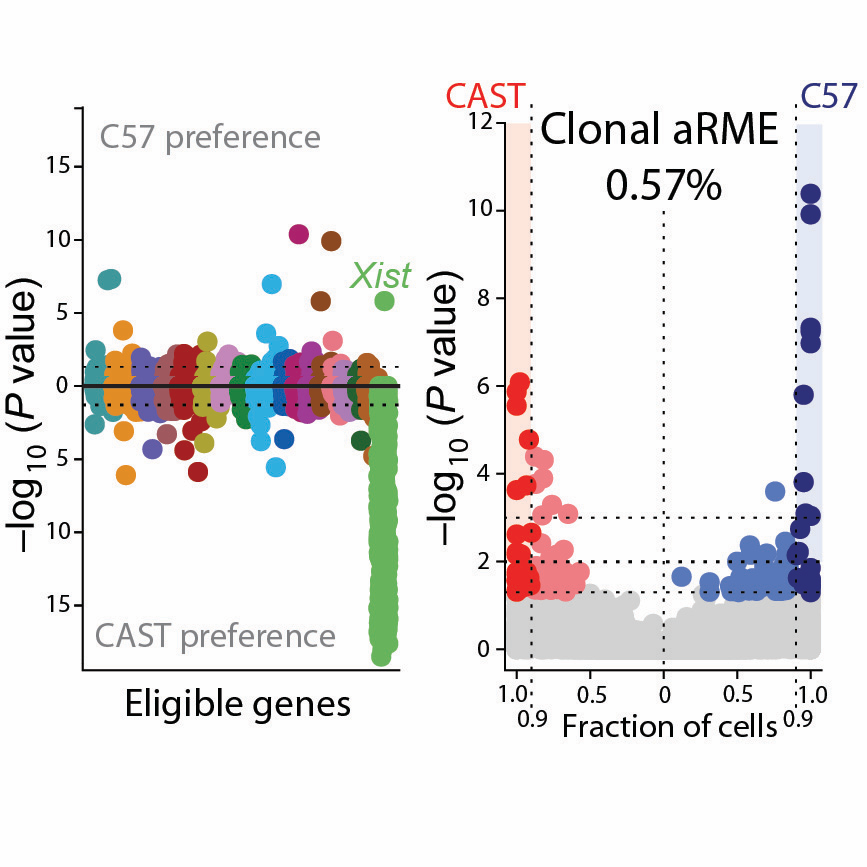
Paper
Analysis of allelic expression in clonal somatic cells
Recent literature have suggested that random monoallelic expression (RME) of autosomal genes is widespread. In this paper, we formally tested whether allelic expression is clonally maintained in somatic cells and we found that clonal RME is scarce, whereas stochastic RME due to transcriptional bursting is widespread.[2016-09-01] [link]

Recognition
Appointed Professor in Molecular Genetics at KI
Rickard has been appointed Professor in Molecular Genetics at Karolinska Institutet, starting Nov 1st 2015.[2015-10-01]

Award
Daniel Ramsköld's PhD thesis awarded 2014
Daniel Ramsköld recieved his Phd in June 2014 and was recently awarded the Dimitris N. Chorafas prize for his contributions to medical research. Congrats Daniel![2015-01-01] [link]