Smart-seq3xpress
We are extremely happy to share our state-of-the-art plate-based method that is cost-efficient, scalable and sensitive!!
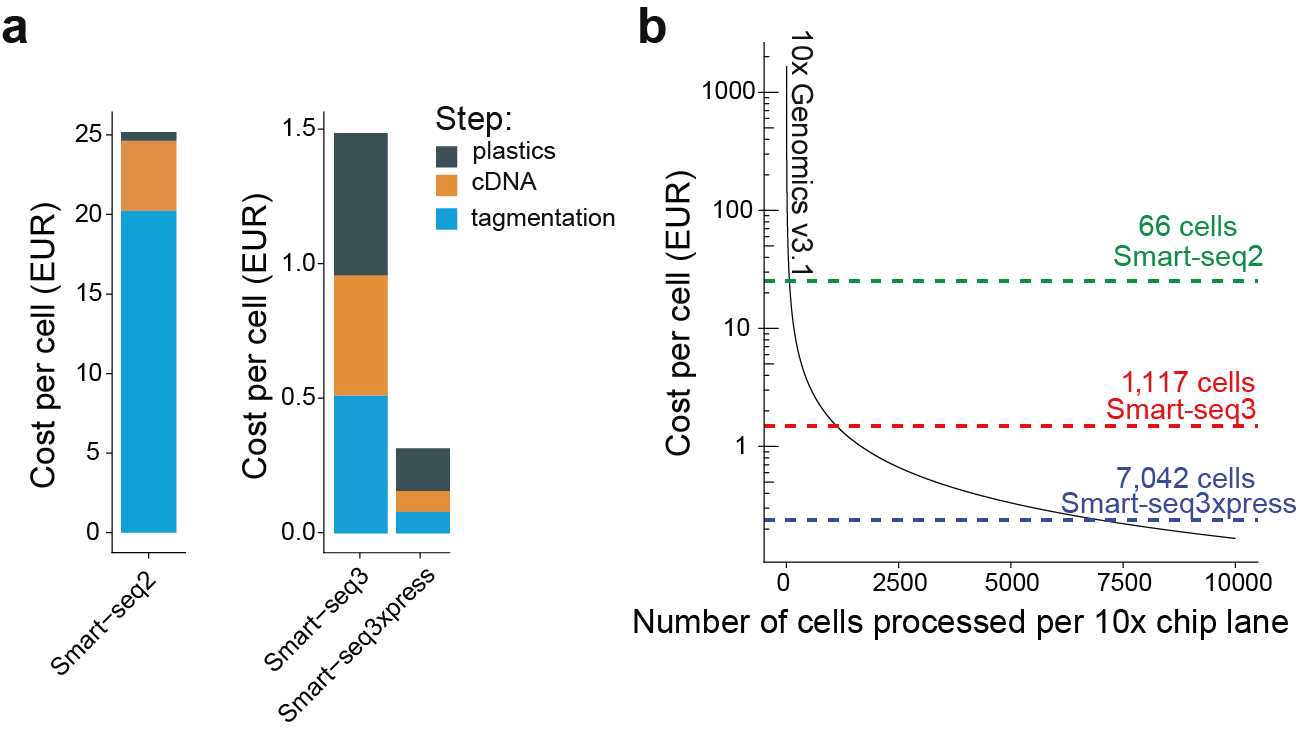
Per Nature Biotechnology publishing rules, we were not able in the publication to state costs per cell, but the below chart demonstrates that cost per cell is now down to a whopping 0.25 EUR with costs per experiments mirroring 10x Genomics libraries.
Given the improved sensitivity, robustness and cost-savings, all users with nanodispensers should definitely move over to Smart-seq3xpress!
Molecular biology protocols:
The most recent protocol for Smart-seq3xpress is available at protocols.io, here.
We will continously link to newer protocol verions, should we make further improvements.
Computational protocols:
We have integrated Smart-seq3 data processing capabilities into zUMIs (Parekh et al. 2018) to correctly parse molecular and cellular barcodes in an efficient analysis pipeline.
The computational pipeline to process Smart-seq3 data, reconstruct RNAs, assign RNAs to alleles and transcript isoforms is similar to Smart-seq3 and available in our github repository smart-seq3.